David J. Patterson–known to friends and colleagues as “Paddy”–is a major figure in eukaryote taxonomy, evolutionary protistology and biodiversity informatics. In the 42 years since his first paper was published (a nifty little study of habituation in Vorticella) he has done enough work for several ordinary careers. With various collaborators, he has added some 250 new taxa to the eukaryote family tree (stramenopiles, Cristamonadida, ramicristates, Centramoebida, among many more), and has had a hand in the early description of a remarkable number of organisms, such as Cafeteria, Carpediemonas, many flavours of Nuclearia and such intriguingly-named creatures as Kamera lens, Macapella, and Massisteria. Anyone doing research on Centrohelida, Nucleariida or heterotrophic flagellates of any kind will find themselves, at some point, following his tracks in the snow.
In the past decade, he has been at the forefront of the effort to put biological diversity online, nudging protistology into the “Big Data World.” An early project in this field was his online database of microbial life, micro*scope, and its sister site bio*pedia. Though the software now rattles and clanks like an old tractor, micro*scope still provides a home for some of the best collections of high-quality, peer-reviewed images of protists on the internet. While preparing this interview, I was delighted to discover that it is once again online and fully interactive at a new address.
I interrogated Paddy by Skype, in late November of 2013, but it was several months before I got up the nerve to pass his very fluent and erudite answers through the microtome. Our conversation ran largely to taxonomy and bioinformatics, and the miseries of microbe identification, which left little time to discuss his four decades in protistology. I hope to remedy that, at some point in the future. In the meantime, here is a fairly thin slice of Paddy.
First, where are you now?
You’re speaking to me in Arizona, at the moment. I’m originally Irish, worked in England for 17 years, and then went to University of Sydney where I became head of Biological Sciences. I have a house there, so that’s where I think is home. For most of the last nine years I’ve been working in the U.S.
In 2004, you went to the Woods Hole Marine Biological Laboratory.
At that time I had an idea that you could use taxonomy as a device in the emerging internet to start joining together information about organisms better than the standard search engines could do, because if you’re playing with taxonomy you understand issues like hierarchies, and the value of hierarchies, and you understand that there are different spellings for organism names and that there are many different names for organisms, different opinions leading to synonymies, vernacular names and so on. Taxonomists are the guys who know all about that stuff, know how to manage it. The idea was to build a digital infrastructure that calls on name so that it can organize all biological information on any species. That was the idea that was the nucleus of the Encyclopedia of Life. I led the informatics team that established the EOL.
You’ve moved on to other things, since then?
Well, I am still in the same vein. Somewhere round about 2000, mostly because of the inaccurate delivery estimates of a shipping company (called Australian Van Lines, not to name anyone in particular), I was unable to write a book that I’d intended to write and my intentions turned more to moving information onto the internet because it was very clear that that would be where people would seek information. Now the process is almost 100 percent complete. It was that need to think of ways to integrate distributed information, because biological data, in our case, is emerging on more than tens of thousands of websites. Each carries fragments of information, some overlapping, some independent. I began to explore how to join the information together and enable the community to contribute, with the micro*scope website. The result was a communal environment that relied on a names-based infrastructure. EOL was the realization of the same idea for all organisms. However, the ideas, tools and services were not available to others, and what has been driving me since then is to promote the development of an infrastructure that is independent of any project. Then we can all use it to combine and filter the distributed data; there is an incentive to collaborate, and to share individual progress. That’s what’s been keeping me busy since EOL.
You’ve been something of a champion of name-based taxonomy, over some of the other options (barcoding, for instance).
I wouldn’t say that. The best approach has to be considered a bit more carefully, because it depends on what the objectives are. Everything that emerges in the world of science has value, and we should never ever dismiss any element.
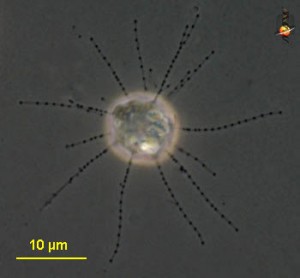
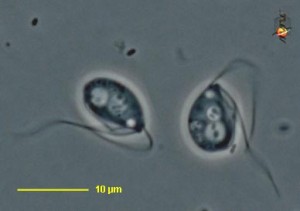
The cercomonad flagellate Massisteria. Image by D. J. Patterson, from micro*scope (click for source)
One of my biggest concerns about science, just thinking about the way it’s grown over the last 200 years or so, has been its faddishness, its tendency to have fashions and for new technologies to knock out older approaches. The shift, coupled with competition for research funding, quickly leads to the argument that older approaches have no longer got any value. The consequence is to go forward with a new technology and dismiss the old ones. Certainly, in the area of alpha taxonomy, observational taxonomy–the kind of thing that you enjoy doing and that I’ve spent a lot of time at–still has an immeasurable amount of value to give in lots of different ways. The observational approach will continue to be a source for discovery. Our observations on diversity are enriched with environmental information, information on the composition of the community, its dynamics, and the various ecological transactions that take place in nature. There is an enlivening value that comes out of watching organisms of all kinds, but especially with microorganisms because you can provide students with an opportunity to get access to many species in a short period of time, in such a small place. Most of us who watch microbes enjoy that experience enormously, and we can watch school kids and students in universities becoming inspired about biology, about the microbial world, about nature through that. So I rate the observational side of things very highly and, yes, I am fearful that the new technologies like the molecular barcoding will simply knock out the old stuff, leaving us with problems, as insights are disconnected from all the old stuff, and we have an impoverished view of Nature.
Clearly, this is not an either/or situation; although competition for funds lead many to project it as if it were. Rather, we have to build bridges. If we don’t invest in finding the technology to create such bridges, then the old stuff is likely to get forgotten, and a new molecular description will take over. If that is not managed, we will have to redescribe all that we know through molecular techniques; and there are invaluable and irreplaceable historical observations that will be lost. The informatics tools and services are emerging, and that will link information together–as long as it is digital and openly accessible [on] the internet. One useful and immediate task is for those who explore diversity through molecular tools to keep voucher specimens, so that they are able to say, “This sequence came from this particular specimen, or this species. I don’t know what the species is, but this is the thing I’ve got the sequence of information from.” And if they can put that onto the internet somewhere, then you and I may be able to say, “Hey, I know what that species is.” Then by just adding the name, the infrastructure will be able to connect the new molecular observations to all of the information about that species that’s available from the time Leeuwenhoek forward.
Looking back over the 300 years since Leeuwenhoek, the effort that’s gone into classification is staggering. The loss of all that work would make a person sad, even if it were not still useful, scientifically.
There is a bit of that that I absolutely agree with. Looking at the way folk tried to sort out all this diversity over a period or 200-250 years helps us a lot to understand fashions in science and the ways things can go wrong. If you’re attuned to the history you can then watch the present and say, “We’re remaking a mistake of this kind.” We should also be able to look at history and not just think about the way that people do things, but start assembling a suite of principles that can guide best practices. One of my biggest concerns about systematic protistology is the failure of good principled approaches to dominate over crappy approaches. And there’s a lot of crap in systematic protistology. A lot. I mean, it’s bad, in my view, terribly bad.
Researching an organism, I often find myself on a moebius strip of identification, because of the insufficient descriptions that are out there…the competing descriptions of the same organisms.
Again, you could build the answer to this. The solution doesn’t exist as a single item. The perspectives you talk about are different points of view. Different folk look at the problem from their own perspective and from their own biases, and so you get many different variations on the theme. Each one usually has something special to add. But also, no document is complete. But, novices especially can’t see the signal for the noise. If you’re in this kind of unclear zone, it is easy to get lost and confused.
The answer to me is simple, and we ought to do it straight away, and it only needs a little bit of money. (Well, probably a fair bit of money!). It relies on accumulating knowledge in little bits, empowering everyone to be part of the process, and having a mechanism by which anyone interested can criticise and evaluate content. The Wikipedia model is great. Begin with a communal classification structure in which the hierarchical component is independent of the terminal taxa, so that you can have multiple classifications of the same species arranged in different ways. That would then allow a user to have a classification like, say, the Adl et al. classification, if that’s what we want, or the Margulis classification, if that’s what you want, or a Cavalier-Smith classification, if that’s what you want, or one that is principled and defensible if that is what you want.
The separation of organizing the classification from the terminal entities gives you flexibility for new insights or different points of view. Once we have a communal catalogue of terminal taxa that allows you to remove all those false Didinium species [Editorial Note: Paddy is referring to six non-existent Didinium species that have spread by “database contagion” to many sites on the internet], we can begin the process of annotating most of those taxa with descriptive information, ideally as what they might call semantically minimal units. Things that would just say: Minimum length? Give a number. Maximum length? Give a number. Minimum width? Give a number. Maximum width? Give a number. Just think of all the descriptive features and find a way to break it down so that you have one-word answers to input. This approach can be accelerated by using defensible hierarchies to annotate all children with the same information (one data entry should annotate all ciliate species with the statement: “Primitively with macronucleus and micronucleus”).
 Another advantage of annotating taxa with atoms of information is that the resulting matrix can be used in a filtering approach to identification. I think we talked about it once using the example of Lucid keys. What Lucid does is to create a grid that includes all the species that the maker knows about and here are all the attributes. To identify the taxon, you simply say, “It’s from freshwater,” and immediately everything that is not known to ever occur in fresh water is eliminated from your system. You can then say, “…and it’s 23 microns long,” and everything that doesn’t have 23 in between minimum and maximum is thrown out as well; and you say, “It’s got 2 flagella and everything that doesn’t have 2 flagella is thrown out. You shorten the list extremely quickly. At some point you can say, “Ach, I can’t go any further, I’ve got no more characters, but we’ve only got seven species left on the list, show me pictures of them.” That would take you to a gallery of those seven species: which one do you think you’ve got?
Another advantage of annotating taxa with atoms of information is that the resulting matrix can be used in a filtering approach to identification. I think we talked about it once using the example of Lucid keys. What Lucid does is to create a grid that includes all the species that the maker knows about and here are all the attributes. To identify the taxon, you simply say, “It’s from freshwater,” and immediately everything that is not known to ever occur in fresh water is eliminated from your system. You can then say, “…and it’s 23 microns long,” and everything that doesn’t have 23 in between minimum and maximum is thrown out as well; and you say, “It’s got 2 flagella and everything that doesn’t have 2 flagella is thrown out. You shorten the list extremely quickly. At some point you can say, “Ach, I can’t go any further, I’ve got no more characters, but we’ve only got seven species left on the list, show me pictures of them.” That would take you to a gallery of those seven species: which one do you think you’ve got?
And building that is elementary, it’s not a challenge in any way to build such a structure. The challenge is to populate it with data. That would take a lot of time. But that’s the kind of thing which a community of protistologists can easily do as a communal on-line endeavour. The current research paradigm, in my view, is never going to build that structure because it’s going to take too long, because it requires resources distributed to a large community of experts, and that’s not the way research works. But a society, or group of enthusiasts could say, “Well, why don’t we make this system and make it as an open and communally available system,” [and] then it could happen.
And something like the micro*scope environment could be adapted to that kind of thing. You already have a classification in there, there’s a place where it can be edited. Micro*scope is the kind of environment that could be used to create such a structure.
For identification purposes, micro*scope is still the most useful site on the web, by a long stretch.
It’s in its death throes, to be honest. Unless somebody very generous comes up with a heap of money that would allow some, what they call, refactoring of the software, it’s not going to survive much longer. Which is a shame. But that reveals a problem with infrastructure: it needs sustained funding. The answer for that probably lies with support from societies or institutions.
Much of the other stuff on the web has not been competently reviewed. There’s a sort of echo effect…when bad information gets out there it reinforces itself.
And it’s often repeated as well, but yes, you’re right. You know I was hunting for something the other day, and what was coming up under image search was just amazing, just so badly identified. There’s a carelessness with so many people, about protistan identification, even by specialists. It’s very worrisome. I have a fairly negative view about the standards that protistologists set themselves; I think they generally set very, very loose standards.
What about the future of morphological identification, in general? In some groups it’s become increasingly difficult to identify organisms, at least with the light microscope.
Yes, there’s always been problems about how much information you can get from a light microscope, and whether it’s appropriate to what you think is the diversity inside any group. If identification is the only thing you’re concerned about, in my view we should just absolutely and enthusiastically embrace various molecular technologies. We should start off by a major effort to get reference material for each taxon sequenced so that we can match new or environmental samples against this reference structure. If, after that, all you want to know is who’s living in a particular place, the best thing to do in the future would be just to put out a sample and mash it up and analyze the total DNA for all the sequences in there.
The trouble is, most don’t want to know a list of names of things that are in that pond. They actually want to see the organisms, they want to watch them. They want to identify after they’ve observed them…and they won’t have access to molecular tools, so you’re back to that world in which microscopes are needed. Again, as before, either/or is not the way to approach this. We need both, and that means, for the molecular community, they need to assemble [quite a large amount of] reference material. And I don’t think it’s that hard to do. It’s just getting a heap of people enthused to work together to populate the environment we will need.
Maybe it would help to enlist amateurs like me, in an organized way, to fill in some of those gaps?
You can take charge of it! And there are, around the world, some very good amateur observers–often more interested in microalgae than protozoa–but they are out there. Societies could endorse their efforts as well, and societies could be looking at training exercises for kids in schools, for example, but that can also add to the infrastructure that we all might use.
Looking from the outside at biodiversity informatics, it seems to me there are a lot of competing projects. How much overlap is there between all the different databases out there?
Before we might talk about some particulars, one thing that we haven’t been doing is to distinguish the conduct of research from the assembly of infrastructure. Bioinformatics is as much an issue of infrastructure as research. Research is typically done with short-term projects that last 3-5 years. Typically, there’s an uncertainty about whether the research will ever be done. People write proposals, and they may or may not get funded, so there’s a kind of uncertainty in there. Typically, in an area, you’re going to have an array of individuals and small groups conducting research.
So, let’s say, in ciliate diversity you’ve got the Foissner group, you’ve got Weibo Song’s group, Denis Lynn…a lot of people. You don’t ever think about those guys as overlapping, of there being any redundancy in there. We don’t want to have a situation where there’s only one person, because then you will have a single point of failure, and you lose the diversity of people and their approaches, something we value in research.
Infrastructure is not the same. During the early phases of development of an infrastructure, it really is very, very good to have diversity. This redundancy should not be condemned, it should be welcomed. That said, I agree there is too much wheel rebuilding within biodiversity informatics infrastructure. The reason is that there is simply no paradigm for funding infrastructure. There’s no mechanism by which we can say, well, what we want to do is to build an infrastructure and it’s got to live in perpetuity, at least as far as we can imagine. It’s going to serve lots of people. Once you have worked out a suitable approach, we need only one infrastructure, but one that’s going to serve lots of research teams all around the world. To do this, we would like to have money for at least the next 10 years — at least. There’s no mechanism to achieve that, currently. So we end up having the situation where everybody argues that they’re building infrastructure but have to present it as innovative research. The result are many competing systems justified [as] being better than the others out there. And then you get duplication. So, the solution is in establishing a different kind of funding paradigm for infrastructure.
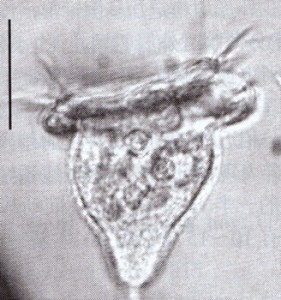
Pseudovorticella pattersoni, one of many species named for Paddy. From micro*scope (click image for source)
In my view, funding agencies have to acknowledge the need for infrastructure for this big data world, to set aside special funds for special funding conditions to allow the infrastructure to be built, with the expectation that the infrastructure is going to be promoted by large consortia of players, ideally all the players in the game, with the peak of funding at the beginning, to get the system in place, but then long-term funding to ensure that it stays there for the long haul.
Ideally, what we would like is, if you publish a paper about a new species of Paramecium (of which there are many, by the way) immediately, the infrastructure gets populated with this information. You would like to see it in a definitive online classification. It should just happen like that! Done. The technology is there, it’s already in place. If you publish a species in the Pensoft series of journals, their workflow captures that information–that’s a new species!–before the paper ever comes out, sends the information over to Zoobank, and the information’s sitting waiting on Zoobank, so the moment that the paper is actually published, a switch is flipped and the information shows in Zoobank. There’s no reason why all journals cannot have this kind of instant publishing system. Once in that digital space, the new information can flow to other environments, such as EOL.
What projects like Catalogue of Life do is to rely on some decrepit expert person. I mean, every taxonomist has a limit to how many taxa they know. Usually it’s around a thousand. Anyone who really knows more than a thousand taxa is remarkable, and even a thousand is doing pretty well, for many. And yet you get these global species databases being created by some individual and they have many, many more than 1000 taxa in them, so it cannot be definitive, they’re going to be wrong. That person is kind of borrowing stuff, guessing what the right status is for these taxa, so you get a sloppy system, you get a system that’s out of date. It doesn’t maintain currency. And the other bit that’s absolutely missing is a mechanism for users to enter in criticisms, or comments or improvements, or corrections. The technology for that also exists, it’s just not being adopted by any of these players. I mean, clearly, what you’d really like is when you go to something like Catalogue of Life and you say what species are there in the genus Paramecium and it comes up with this little list of 6 or 7 species and I know that there are at least 28 good species in Paramecium, I would like to say, well, what about…and then run the list of them, or feed them in, but their architecture, their workflow, just doesn’t permit that. And that way it stays bad, and so…
[interrupting] Your vision for EOL was that it would incorporate that sort of feedback from users…
Yes, and they achieved a system a bit like that, but not exactly the way I wanted. Certainly, when I was thinking about that it wasn’t in the normal style of the contemporary systems.
So, the ideal thing to minimize duplication is that you identify uniquely every piece of information that’s made available…doesn’t matter where it is or what it is. So, if you have a number that’s long enough you can make an index number that’s going to cover everything that’s ever going to happen in the world. These big (32 digit) numbers are called “universal unique identifiers” (UUIDs), so every statement everywhere, every piece of information can have one of these things attached to it.
The solution to data quality improvement in my view is annotation, which is a technology that’s been around for some time. It’s well in place in some areas, but not in biology. And what annotation does is it monitors things with those UUIDs, and then provides a little kind of plug-in to your computer, so if you’re reading something with your browser every single item, such as a name in COL, is distinguished with its own UUID. If we see something such as a misspelling you can click on it, using your little plugin, and out will come a commentary panel, probably a drop-down menu, and you say, “Misspelling?” and you submit. Now, that comment, that annotation, is attached to that UUID, which is attached to a piece of data. That should flow back to where that piece of information first came from, so Catalogue of Life should suddenly get a message popup saying, “Ah, Brucie over there, he’s just decided that there’s an error in this. You should check this out.” It also means that anybody else who looks at the same data should also be able to have a panel where they can see any comments. So they’d know that there may be doubts about this. But in principle, you could start putting annotations against all of those misidentifications, or soft identifications, saying “I’m not sure these guys have identified this thing right. It doesn’t gel with the thing I’ve seen over in that website.” The technology to build that kind of system is there.
Are there enough young taxonomists being produced by the universities. Are they being turned out in sufficient number to handle the load?
You shouldn’t have asked the “handle the load” bit of the question, you should have just stopped with the first bit, because that takes you into the area of what “load” is. I think the answer is “No, there is not.” You can understand the problem with universities, that they must justify themselves by preparing students for the work force, and the students need to feel confident that they’ve got an array of skills that are going to position them to be competitive within the work force. And so, if we train them in out-of-date areas that the workplace doesn’t require, like looking at protozoa down a microscope, then we’re not serving the students well, and if we’re not serving the students well the universities are not going to get applause, and they’re going to go backwards.
So, there’s a lot of pressure to keep moving forwards in using all the new technologies. But I honestly cannot see how you can say you have properly trained a biologist if you don’t build skills in looking at organisms, because it’s only through knowing the organisms and watching them that you know what questions to start asking. And if you don’t have that bit of the game, then somehow you’ve become a charlatan. You’re picking up someone else’s questions and using them. You’ve become just a workman, no longer a visionary.
But then you said “load,” and the question becomes what might “load” be? And I don’t know how I would answer that. I think these days the general sense is that the load that taxonomists have to carry is getting smaller because people don’t seem to want taxonomists very much, so how much load do they have to carry?
Why? What has changed?
I think the promise of the molecular technologies has led to serious questions as to whether traditional taxonomy is an appropriate way of recording biodiversity.
Somebody used the word “quaint” in an article I was reading, suggesting that there was something outmoded about it.
Yes, there’s a lot in that. I think a lot of the alpha taxonomists can be fairly irresponsible in terms of how much energy and time they’re willing to invest in a very small problem. But that also takes you into the fact that taxonomy is actually very expensive to do. Very expensive. Because good taxonomy relies on people with a lot of experience, so you’re going up in the salary level and you need to invest a lot of time. I mean, you imagine how much time it takes to get a good identification of a single species, especially if it’s in one of the nasty genera.
Never mind revise an entire group...
Or provide a catalogue for what’s in a pond.
You have done a lot of popular outreach, with books aimed at a general readership, like Free-living Freshwater Protozoa, and Seen and Unseen: Discovering the microbes of Yellowstone. What prompted you to write for the public?
Well, it came out of being a university teacher. It’s just part that environment. What was very obvious to me as a teacher was that in trying to teach people about protozoa, or a protozoan perspective on things. There are many topics that are greatly enriched by taking a microbial or protistan perspective, cell biology being an outstanding example, because if you want to find what cells have got and what they can do with it, you go look at protist cells, you don’t get as good insights by looking at mammal cells or anything like that. Ecology, trying to understand ecosystems and the general rules of ecosystems: most of them break when you go down to the tiny guys, because there are so many different factors at play, and you don’t have a reasonable and fair grasp of ecology unless you can cover the full spectrum.
But yet, trying to teach people these things in university, I found I was at a massive disadvantage over those who were teaching about, say, flowering plants or birds, because everybody knows what a bird looks like and they know how they behave and they know much of their biology just from common knowledge; ditto plants, ditto larger organisms. And yet when you start trying to tell people about protists, they have no idea what kind of organisms you’re talking about, no idea of where they live, how they live, what all the terms are, so you have to go back a lot earlier in the educational process. Plus, I was watching others dealing with the same problem that you were talking about at the beginning, which is, bad identification. Both of those issues can be addressed by trying to introduce a microbial perspective, or a protistan perspective, into peoples’ awareness at a much earlier stage than we currently do. And there were things like the color atlas book, there was another book that was for prawn farms, oddly enough–because they’re kind of interesting environments, microbially speaking–sewage farms are always very interesting.
So, from the books then came micro*scope, which again has the same kind of purpose, but that one, the thing that was driving me for that was partly to get knowledge onto the internet because I came to the conclusion that in the future–this was in 1999, 2000–people were not going to go to books for their information, they were going to go to the internet. So, the intent was to get as much as possible into websites. But a second big driver for micro*scope was actually the poor quality of the micrographs that were then available on the internet.
In those days, the micrographs were just absolutely terrible. Well, you’d have the misidentification, you’d have bright fields, you wouldn’t have any contrast, you’d have no reference to the defining features of the organisms. You wouldn’t see the things you really needed to see, to know that it was what it was. You’d have dust and other junk over it, you’d get two thirds of the organisms, because it moved when the photograph was being taken, and its tail’s off the end of the picture. Or they were fixed, which was not at all helpful if you were working with living material. So, one of the drivers behind micro*scope was to make the sure that the site was made a rich and rewarding place to go into and look around, because the pictures were strong visually.
Where did you get the name Paddy?
In Northern Ireland, adding a “Y” onto somebody’s name is the normal way of creating a diminutive, so William is usually Billy, James is Jamesy, Jim is Jimmy, and with a name like Patterson it was the Patterson bit that got the diminutive, so it was shortened to Paddy. My sister my brother and I were all called Paddy, so when somebody would ring the house and ask for Paddy they’d have to work out which one they were talking about. I’m the only one who hung onto it; but I hung onto it because Paddy is also the word English people use for Irishmen, and to call someone a Paddy is to declare them to be stupid and definitely a lesser form of life than a human being, let alone an English human being: a very derogatory stance that the English take to Irish generally (or to the rest of the world, generally, in fact. So, my first day at University of Bristol after being appointed to be a lecturer, I was going around to introduce myself to various people. I came up to one room and the door was open, and there were two guys talking in there, and one of them said, “I hear we’ve just appointed a Paddy as a new lecturer.” And so I knocked on the door and I walked in and said “Hi, I’m Paddy.” Since then it’s stuck, professionally as well as privately.
I want to ask about a genus that was named after you.
Pattersoniella vitiphila?
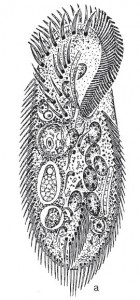
Pattersoniella vitiphila (drawing by Wilhelm Foissner, from Helmut Berger’s Monograph of the Oxytrichidae, 1999) Click Image for link to the book.
Yes, in one of the descriptions on EOL it’s described as “A real cutey, from Fiji.”
Vitiphila is the wrong name, it should actually be “vitiensis.” “Vitiensis” would mean, “…of Fiji.” So, way, way back I heard that the Royal Society of London had been donated money in the 19th and early 20th centuries to help scientists to carry out field work. By the time we got to the 1970s and 80s, nobody really did much field work and the money wasn’t being used. So I wrote to the Royal Society and said, do you know, I have this thing that I think is important to work on, and that is to work out whether protozoa have biogeography. And the way I think we should do that is to compare a list of species that one finds one place that has been well studied, such as Bristol, which is where I was, with somewhere that has hardly been studied at all, and if we find the same species in both places then we’ve got cosmopolitanism and if we have that are very different then we’ve got endemism, and the place that I recommend you send me to, with my microscope, is Fiji. And they said “Yes.” So they sent me to Fiji, and then that began a whole series of studies on biogeography. That ciliate was found during that trip to Fiji, sent to Willi [Foissner] and I asked him to use the name “vitiphila” which is “likes Fiji” rather than “of Fiji.”
As long as we’re talking about protist biogeography, where do you stand on that?
The answer will depend on the level of resolution that you wish to apply. Obviously, at any level everything has got an endemism. I mean, you are an entity and you have very limited distribution. I am an entity and I have a distribution that barely overlaps with yours; but Homo sapiens is a different entity: cruder resolution, worldwide distribution. At the level of biodiversity that interests me, which is morphological distinctiveness, the vast majority of protists are cosmopolitan. There are going to be a few that are not, but we found things like Postgaardi, which is a flagellate, down near the bottom of euglenozoan territory, which lives in very, very strange anoxic, hypersaline environments, and it was originally found somewhere up in the Arctic circle; and the next time it’s found it’s somewhere in Australia; then it’s found somewhere in the Antarctic. When you’ve only got the one record you’re thinking, well, this could be endemic, and as time goes by, more observations are made, and you pick it up elsewhere. Endemism changes to cosmopolitanism.
I mean, people would send me bottles of water, at one stage, when I would doing this microbial biodiversity stuff for marine ecosystems…they would say, “We’ve just been taking water samples, and there’s a flagellate in here that’s in vast numbers, and it’s obviously got big, important influence on the ecosystem, and we’d like somebody to identify it, and we don’t know how to identify it, so we’re sending it to you, could you identify it, because you’re a specialist.” And I would say, yes, it’s Cafeteria roenbergensis, and I would tell them that without opening the bottle, and I was almost invariably right. Because it’s the most common thing, and when you collect something and you grow it for a while, the weeds dominate and knock everything else out, and Cafeteria‘s one of them.
You should be asking me questions about names like “Cafeteria“…and “Massisteria.”
And “Carpediemonas“…
That was named after my wife who was another protistologist, but she died in ’94. That was after her, because her motto, in life, was “carpe diem.” Carpediemonas was one of the ones she had worked on. And then Alastair [Simpson] came up with Ergobibamus, “So, let us drink.” And we wanted to create another one for Gaudeamus, “and be happy,” but Gaudeamus has been picked up by somebody else for some kind of fossil mole, or some piece of junk like that. I enjoy names that are sonorous and amusing.
What is some of the work you’re most happy to have done?
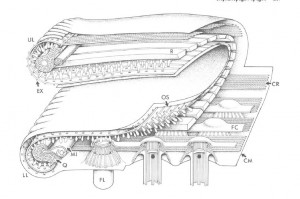
Diagram of Stephanopogon cortical ultrastructure in the mouth region, by D. J. Patterson. Click image for Source.
There was an approach to evolutionary systematics that I really valued that came up as a kind of variant form of cladistics and that was called transformed cladistics, and it gave greatest emphasis to the evolutionary novelties that defined new branches in the tree of life. And I held then, when I started learning about that in the eighties–and still do–that it was almost certainly the most sensible way forward. And there was a heap of papers where I started to explore issues like that and to criticize other approaches to evolutionary analyses. There’s a paper on Opalina, that I did, in which is a little tree and it looks very, very simple at the back but it’s one of these ones that…here’s the tree and here are the events that define each of the lines of the tree. That to me was the way things ought to be done, I still think it’s the way it ought to be done. I came up with some new group names, including Slopalinida — which is one of the names I wish I hadn’t come up with–but it was sensu lato Opalinida, so it took the Opalinida and expanded it. The stramenopiles paper is always something that I think was a very good one. There’s a paper with a guy called Robin Smith, where we compared different ways of analyzing data on heliozoa. We showed that A) no algorithm gave a better result than any other one, and B) what emerges from all of the analysis as a kind of consensus we knew anyway so a lot of subsequent analysis is just a waste of time. It’s always nice to say that, yeah, you’re all wasting your time! The Stephanopogon paper is one that I like a lot. I liked the paper, and I liked doing the drawing of the mouth.
Do you enjoy drawing?
I used to, until the day I sat down, put my pen to the paper and couldn’t see the tip of the pen. That was annoying. That was the day my eyes decided they’d got too old to work properly.


Bruce,
thank you for posting this interview – i am particularly interested in his views on EOL /COL and micro*scope. it is a pity that he is pessimistic about the future of micro*scope.
Brian
Well, maybe some funding will become available for “refactoring” of micro*scope’s code. In the meantime, it’s still chugging along at the new address, and is once again accepting edits and uploads.
Hi Bruce,
This is a very very interesting interview. Thanks!
Ferry
Hi Ferry.
Nice to hear that Paddy is still active and enjoying protists.
Thank you for this interview.